#include <solver.h>
Public Member Functions | |
| Solver (PSPM_SolverType _method, std::string ode_method="rk45ck") | |
| void | addSystemVariables (int _s) |
| void | addSpecies (int _J, double _xb, double _xm, bool log_breaks, Species_Base *_mod, int n_extra_vars, double input_birth_flux=-1) |
| void | addSpecies (std::vector< double > xbreaks, Species_Base *_mod, int n_extra_vars, double input_birth_flux=-1) |
| int | n_species () |
| void | setEnvironment (EnvironmentBase *_env) |
| void | resetState (double t0=0) |
| void | resizeStateFromSpecies () |
| void | initialize () |
| void | copyStateToCohorts (std::vector< double >::iterator state_begin) |
| void | copyCohortsToState () |
| double | maxSize () |
| void | updateEnv (double t, std::vector< double >::iterator S, std::vector< double >::iterator dSdt) |
| void | calcRates_FMU (double t, std::vector< double >::iterator S, std::vector< double >::iterator dSdt) |
| void | calcRates_iFMU (double t, std::vector< double >::iterator S, std::vector< double >::iterator dSdt) |
| void | stepU_iFMU (double t, std::vector< double > &S, std::vector< double > &dSdt, double dt) |
| void | calcRates_EBT (double t, std::vector< double >::iterator S, std::vector< double >::iterator dSdt) |
| void | addCohort_EBT () |
| void | removeDeadCohorts_EBT () |
| void | calcRates_CM (double t, std::vector< double >::iterator S, std::vector< double >::iterator dSdt) |
| void | addCohort_CM () |
| void | removeCohort_CM () |
| template<typename AfterStepFunc > | |
| void | step_to (double tstop, AfterStepFunc &afterStep_user) |
| void | step_to (double tstop) |
| double | calcSpeciesBirthFlux (int k, double t) |
| std::vector< double > | newborns_out (double t) |
| std::vector< double > | u0_out (double t) |
| void | print () |
| template<typename wFunc > | |
| double | integrate_x (wFunc w, double t, int species_id) |
| Computes the integral \[I = \int_{x_b}^{x_m} w(z,t)u(z)dz\] for the specified species. For details, see integrate_wudx_above. More... | |
| template<typename wFunc > | |
| double | integrate_wudx_above (wFunc w, double t, double xlow, int species_id) |
| Computes the partial integral \[I = \int_{x_{low}}^{x_m} w(z,t)u(z)dz\] for the specified species. More... | |
| std::vector< double > | getDensitySpecies_EBT (int k, std::vector< double > breaks) |
Public Attributes | |
| OdeSolver | odeStepper |
| EnvironmentBase * | env |
| double | current_time |
| std::vector< double > | state |
| std::vector< double > | rates |
| std::vector< Species_Base * > | species_vec |
| struct { | |
| double ode_eps = 1e-6 | |
| double ode_initial_step_size = 1e-6 | |
| double convergence_eps = 1e-6 | |
| double cm_grad_dx = 1e-6 | |
| bool update_cohorts = true | |
| int max_cohorts = 500 | |
| double ebt_ucut = 1e-10 | |
| double ebt_grad_dx = 1e-6 | |
| double ode_rk4_stepsize = 0.1 | |
| double ode_ifmu_stepsize = 0.1 | |
| bool ifmu_centered_grids = true | |
| bool integral_interpolate = true | |
| } | control |
| bool | use_log_densities = true |
Private Attributes | |
| PSPM_SolverType | method |
| int | n_statevars_internal = 0 |
| int | n_statevars_system = 0 |
Detailed Description
Constructor & Destructor Documentation
◆ Solver()
| Solver::Solver | ( | PSPM_SolverType | _method, |
| std::string | ode_method = "rk45ck" |
||
| ) |
Definition at line 37 of file solver.cpp.
Member Function Documentation
◆ addCohort_CM()
| void Solver::addCohort_CM | ( | ) |
Definition at line 58 of file cm.cpp.
◆ addCohort_EBT()
| void Solver::addCohort_EBT | ( | ) |
Definition at line 70 of file ebt.cpp.
◆ addSpecies() [1/2]
| void Solver::addSpecies | ( | int | _J, |
| double | _xb, | ||
| double | _xm, | ||
| bool | log_breaks, | ||
| Species_Base * | _mod, | ||
| int | n_extra_vars, | ||
| double | input_birth_flux = -1 |
||
| ) |
Definition at line 106 of file solver.cpp.
◆ addSpecies() [2/2]
| void Solver::addSpecies | ( | std::vector< double > | xbreaks, |
| Species_Base * | _mod, | ||
| int | n_extra_vars, | ||
| double | input_birth_flux = -1 |
||
| ) |
Definition at line 42 of file solver.cpp.
◆ addSystemVariables()
| void Solver::addSystemVariables | ( | int | _s | ) |
Definition at line 116 of file solver.cpp.
◆ calcRates_CM()
| void Solver::calcRates_CM | ( | double | t, |
| std::vector< double >::iterator | S, | ||
| std::vector< double >::iterator | dSdt | ||
| ) |
Definition at line 9 of file cm.cpp.
◆ calcRates_EBT()
| void Solver::calcRates_EBT | ( | double | t, |
| std::vector< double >::iterator | S, | ||
| std::vector< double >::iterator | dSdt | ||
| ) |
Definition at line 8 of file ebt.cpp.
◆ calcRates_FMU()
| void Solver::calcRates_FMU | ( | double | t, |
| std::vector< double >::iterator | S, | ||
| std::vector< double >::iterator | dSdt | ||
| ) |
Definition at line 11 of file fmu.cpp.
◆ calcRates_iFMU()
| void Solver::calcRates_iFMU | ( | double | t, |
| std::vector< double >::iterator | S, | ||
| std::vector< double >::iterator | dSdt | ||
| ) |
◆ calcSpeciesBirthFlux()
| double Solver::calcSpeciesBirthFlux | ( | int | k, |
| double | t | ||
| ) |
Definition at line 352 of file solver.cpp.
◆ copyCohortsToState()
| void Solver::copyCohortsToState | ( | ) |
Definition at line 297 of file solver.cpp.
◆ copyStateToCohorts()
| void Solver::copyStateToCohorts | ( | std::vector< double >::iterator | state_begin | ) |
Definition at line 256 of file solver.cpp.
◆ getDensitySpecies_EBT()
| std::vector< double > Solver::getDensitySpecies_EBT | ( | int | k, |
| std::vector< double > | breaks | ||
| ) |
Definition at line 430 of file solver.cpp.
◆ initialize()
| void Solver::initialize | ( | ) |
Definition at line 194 of file solver.cpp.
◆ integrate_wudx_above()
| double Solver::integrate_wudx_above | ( | wFunc | w, |
| double | t, | ||
| double | xlow, | ||
| int | species_id | ||
| ) |
Computes the partial integral
\[I = \int_{x_{low}}^{x_m} w(z,t)u(z)dz\]
for the specified species.
- Parameters
-
w A function or function-object of the form w(int i, double t)that returns a double. It can be a lambda. This function should access the 'i'th cohort and compute the weight from the cohort's properties. The functionwshould be able access to theSolverin order to access cohorts.t The current time (corresponding to the current physiological state). This will be passed to wto allow the weights to be a direct function of time.xlow The lower limit of the integral species_id The id of the species for which the integral should be computed
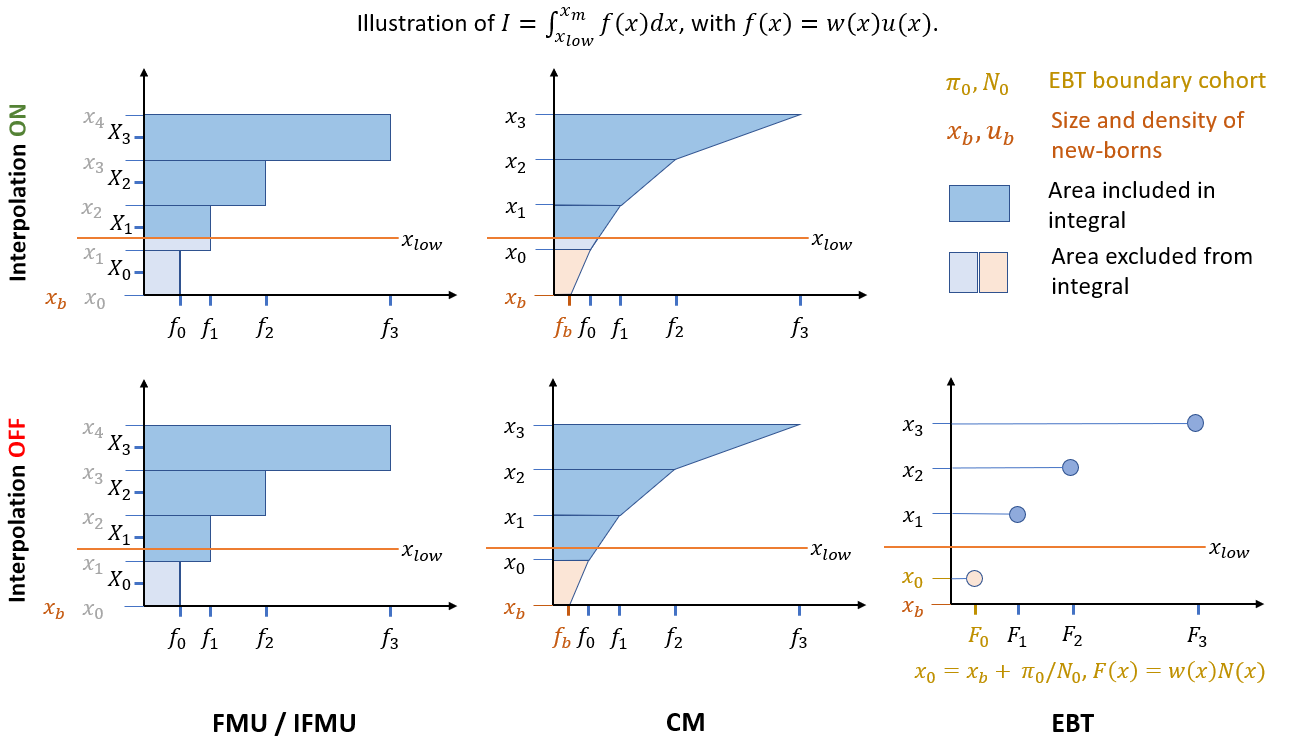
the computation of this integral depends on the solver method. For different solvers, the integral is defined as follows:
FMU: \(\quad I = \sum_{i=i_0}^J h_i w_i u_i\)
EBT: \(\quad I = \sum_{i=i_0}^J w_i N_i\), with \(x_0 = x_b + \pi_0/N_0\)
CM : \(\quad I = \sum_{i=i_0}^J h_i (w_{i+1}u_{i+1}+w_i u_i)/2\)
where \(i_0 = \text{argmax}(x_i \le x_{low})\), \(h_i = x_{i+1}-x_i\), and \(w_i = w(x_i)\).
If interpolation is turned on, \(h_{i_0}=x_{i_0+1}-x_{low}\), whereas \(u(x_{low})\) is set to \(u(x_{i_0})\) in FMU and calculated by bilinear interpolation in CM (See Figure). Interpolation does not play a role in EBT.
In the CM method, the density of the boundary cohort is obtained from the boundary condition of the PDE: \(u_b=B/g(x_b)\), where \(B\) is the input flux of newborns. In the current implementation of the CM method, \(B\) must be set to a constant. In future implementations which may allow real-time calculation of \(B\), \(u_b\) must be calculated recurvisely by solving \(u_b = B(u_b)/g(x_b)\), where \(B(u_b) = \int_{x_b}^{x_m} f(x)u(x)dx\).
Definition at line 44 of file size_integrals.tpp.
◆ integrate_x()
| double Solver::integrate_x | ( | wFunc | w, |
| double | t, | ||
| int | species_id | ||
| ) |
Computes the integral
\[I = \int_{x_b}^{x_m} w(z,t)u(z)dz\]
for the specified species. For details, see integrate_wudx_above.
Definition at line 156 of file size_integrals.tpp.
◆ maxSize()
| double Solver::maxSize | ( | ) |
Definition at line 158 of file solver.cpp.
◆ n_species()
| int Solver::n_species | ( | ) |
Definition at line 153 of file solver.cpp.
◆ newborns_out()
| vector< double > Solver::newborns_out | ( | double | t | ) |
Definition at line 364 of file solver.cpp.
◆ print()
| void Solver::print | ( | ) |
Definition at line 167 of file solver.cpp.
◆ removeCohort_CM()
| void Solver::removeCohort_CM | ( | ) |
◆ removeDeadCohorts_EBT()
| void Solver::removeDeadCohorts_EBT | ( | ) |
◆ resetState()
| void Solver::resetState | ( | double | t0 = 0 | ) |
Definition at line 123 of file solver.cpp.
◆ resizeStateFromSpecies()
| void Solver::resizeStateFromSpecies | ( | ) |
Definition at line 132 of file solver.cpp.
◆ setEnvironment()
| void Solver::setEnvironment | ( | EnvironmentBase * | _env | ) |
Definition at line 143 of file solver.cpp.
◆ step_to() [1/2]
| void Solver::step_to | ( | double | tstop, |
| AfterStepFunc & | afterStep_user | ||
| ) |
- Parameters
-
tstop The time until which the ODE solver should be stepped. The stepper will stop exactly at tstop, for which the final step size is truncated if necessary. afterStep_user A function of the form f(double t)to be called after every successful ODE step.
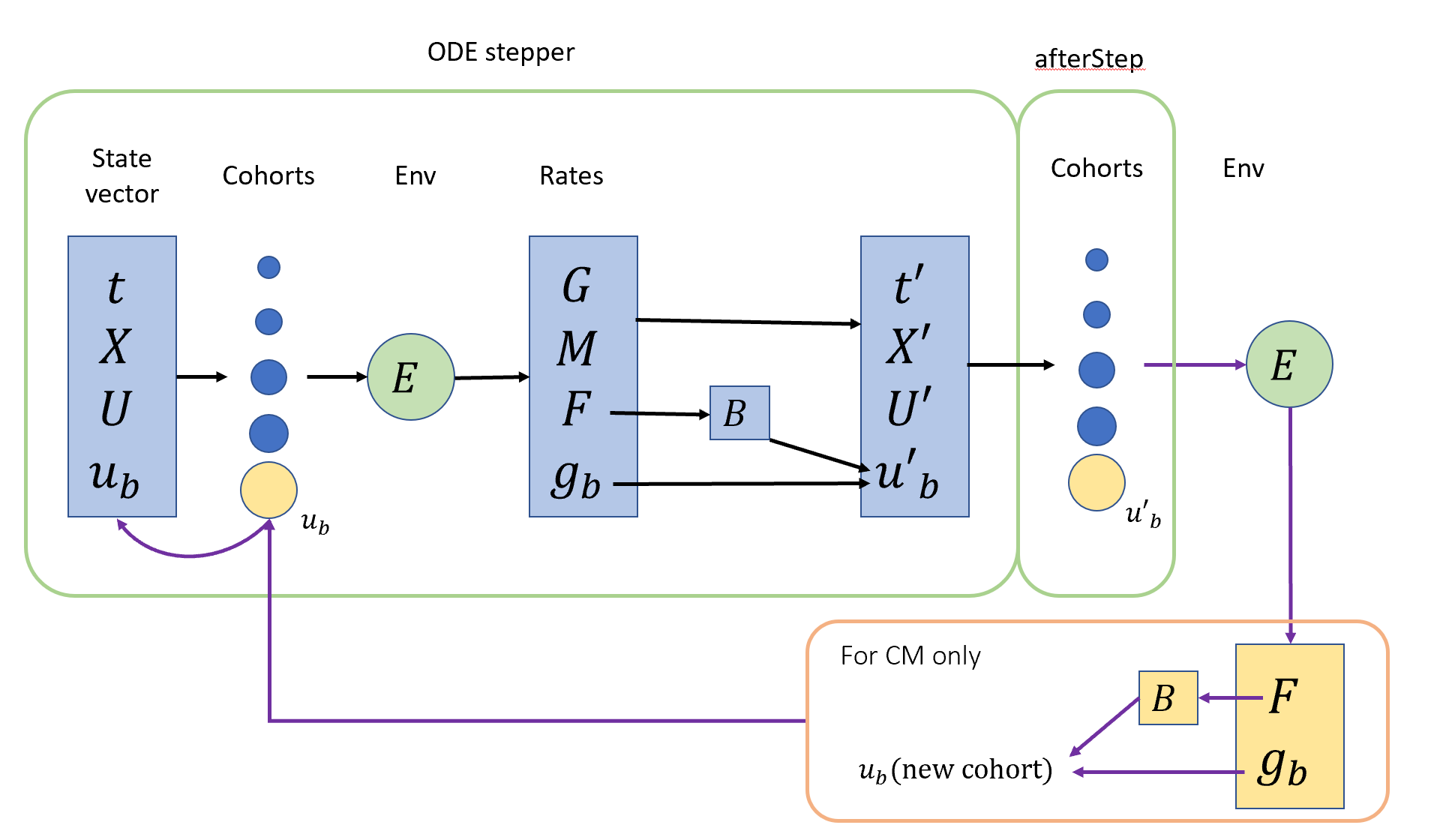
- Note
- 1.
current_timeis updated by the ODE solver at every (internal) step -
2. After the last ODE step, the state vector is updated but cohorts still hold an intenal ODE state (y+k5*h etc). normally, this will not be a problem because state will be copied to cohorts before rates call of the next timestep. But since add/remove cohort after step_to will rewrite the state from cohorts, the updated state vector will be lost. To avoid this, we should ensure that the state is copied to cohorts after every successful ODE step (or at least after completion of
step_to). This is achieved by the afterStep function
Definition at line 15 of file solver.tpp.
◆ step_to() [2/2]
| void Solver::step_to | ( | double | tstop | ) |
◆ stepU_iFMU()
| void Solver::stepU_iFMU | ( | double | t, |
| std::vector< double > & | S, | ||
| std::vector< double > & | dSdt, | ||
| double | dt | ||
| ) |
Definition at line 25 of file ifmu.cpp.
◆ u0_out()
| vector< double > Solver::u0_out | ( | double | t | ) |
Definition at line 391 of file solver.cpp.
◆ updateEnv()
| void Solver::updateEnv | ( | double | t, |
| std::vector< double >::iterator | S, | ||
| std::vector< double >::iterator | dSdt | ||
| ) |
Definition at line 344 of file solver.cpp.
Member Data Documentation
◆ cm_grad_dx
◆ control
| struct { ... } Solver::control |
◆ convergence_eps
◆ current_time
◆ ebt_grad_dx
◆ ebt_ucut
◆ env
| EnvironmentBase* Solver::env |
◆ ifmu_centered_grids
◆ integral_interpolate
◆ max_cohorts
◆ method
|
private |
◆ n_statevars_internal
◆ n_statevars_system
◆ ode_eps
◆ ode_ifmu_stepsize
◆ ode_initial_step_size
◆ ode_rk4_stepsize
◆ odeStepper
◆ rates
◆ species_vec
| std::vector<Species_Base*> Solver::species_vec |
◆ state
◆ update_cohorts
◆ use_log_densities
The documentation for this class was generated from the following files:
- include/solver.h
- src/cm.cpp
- src/ebt.cpp
- src/fmu.cpp
- src/ifmu.cpp
- src/size_integrals.tpp
- src/solver.cpp
- src/solver.tpp
